Documentation > Explore
Your model inputs can be sampled in the traditional way, by using a grid (or regular) sampling, or by sampling uniformly inside their respective domains. For higher dimension input space, specific statistics techniques ensuring low discrepancy like Latin Hypercube Sampling and SobolSequence are available.
You can also use your own DoE in OpenMOLE, by providing a CSV file containing your samples to OpenMOLE.
The
The arguments of the
The
For more details about hooks, check the corresponding Language page.
The
The arguments for
There are some arguments specific to the
For more details about hooks, check the corresponding Language page.
Content:
Execute a Single Run 🔗
Before exploring you model your model you might want to run it for an single set of inputs. To achieve it, the syntax is the following:val input_i = Val[Int]
val input_j = Val[Double]
SingleRun(
evaluation = my_own_model,
input = Seq(
input_i := 10,
input_j := 10.0)
) hook display
Design of Experiment 🔗
Design of Experiment (DoE) is the art of setting up an experimentation. In a model simulation context, it boils down to declaring the inputs under study (most of the time, they're parameters) and the values they will take, for a batch of several simulations, with the idea of revealing a property of the model (e.g. sensitivity).Your model inputs can be sampled in the traditional way, by using a grid (or regular) sampling, or by sampling uniformly inside their respective domains. For higher dimension input space, specific statistics techniques ensuring low discrepancy like Latin Hypercube Sampling and SobolSequence are available.
You can also use your own DoE in OpenMOLE, by providing a CSV file containing your samples to OpenMOLE.
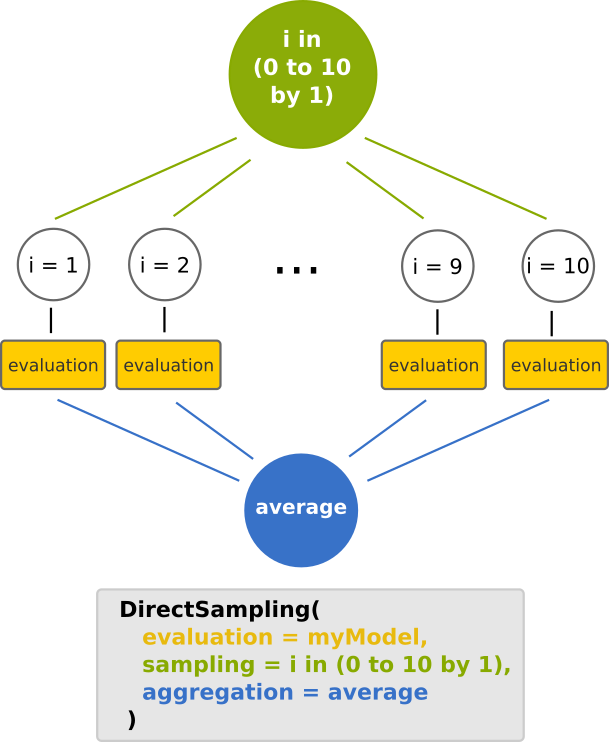
The DirectSampling method 🔗
In OpenMOLE, a DoE is set up through theDirectSampling constructor.
This constructor will generate a workflow, which is illustrated below.
You may recognize the map reduce design pattern, provided that an aggregation operator is defined (otherwise it would just be a map :-) )

Sampling over several inputs 🔗
Samplings can be performed over several inputs domains as well as on several input types, using the cartesian product operator x as follow:val i = Val[Int]
val j = Val[Double]
val k = Val[String]
val l = Val[Long]
val m = Val[File]
val b = Val[Boolean]
DirectSampling(
evaluation = myModel,
sampling =
(i in (0 to 10 by 2)) x
(j in (0.0 to 5.0 by 0.5)) x
(k in List("Leonardo", "Donatello", "Raphaël", "Michelangelo")) x
(l in RandomSequence[Long](size = 10)) x
(m in (workDirectory / "dir").files.filter(f => f.getName.startsWith("exp") && f.getName.endsWith(".csv"))) x
(b in TrueFalse)
) hook(workDirectory / "path/of/a/file")
The
DirectSampling task executes the model myModel for every possible combination of the 5 inputs provided in the sampling parameter.
The hook provided after the task will save the results of your sampling in a file, see the next section for more details about this hook.
The arguments of the
DirectSampling task are the following:
evaluationis the task (or a composition of tasks) that uses your inputs, typically your model task,samplingis where you define your DoE, i.e. the inputs you want varied,aggregation(optional) is an aggregation operation to be performed on the outputs of your evaluation task.
l parameter is a uniform sampling of 10 numbers of the Long type, taken in the [Long.MIN_VALUE; Long.MAX_VALUE] domain of the Long native type.
More details can be found here.
The
m parameter is a sampling over different files that have been uploaded to the @b{workDirectory}.
The files are explored as items of a list, gathered by the files() function and applied on the dir directory.
Optionally, this list of files can be filtered with any String => Boolean functions such as contains(), startswith(), endswith() (see the Java Class String Documentation for more details.
More information on this sampling type here.
Hook 🔗
The @code{hook} keyword is used to save or display results generated during the execution of a workflow. The generic way to use it is to write either @code{hook(workDirectory / "path/of/a/file")} to save the results in a file, or @code{hook display} to display the results in the standard output. @br@br There are some arguments specific to theDirectSampling method which can be added to the hook:
outputis to choose what to do with the results as shown above, either a file path or the word @code{display},values = Seq(i, j)specifies which variables from the data flow should be saved or displayed, by default all variables from the dataflow are used,
val i = Val[Int]
val j = Val[Double]
DirectSampling(
evaluation = myModel,
sampling =
(i in (0 to 10 by 2)) x
(j in (0.0 to 5.0 by 0.5))
) hook(
output = display,
values = Seq(i)
)
For more details about hooks, check the corresponding Language page.
Model replication 🔗
If your model is stochastic, you may want to define a replication task to run several replications of the model for the same parameter values. This is similar to using a uniform distribution sampling on the seed of the model, and OpenMOLE provides a specific constructor for this, theReplication task.
The
Replication sampling is used as follow:
val mySeed = Val[Int]
val i = Val[Int]
val o = Val[Double]
val myModel =
ScalaTask("val rng = Random(mySeed); val o = i * 2 + 0.1 * rng.nextDouble()") set (
inputs += (i, mySeed),
outputs += (i, o)
)
Replication(
evaluation = myModel,
seed = mySeed,
sample = 100,
aggregation = Seq(o evaluate median)
) hook display
The arguments for
Replication are the following:
evaluationis the task (or a composition of tasks) that uses your inputs, typically your model task and a hook.seedis the prototype for the seed, which will be sampled with an uniform distribution in its domain (it must ba a Val[Int] or a Val[Long]). This prototype will be provided as an input to the model.sample(Int) is the number of replications.index(optional, Val[Int]) is an optional variable Val that can provide you with a replication index for each replication.distributionSeed(optional, Long) is an optional seed to be given to he uniform distribution of the seed (\"meta-seed\"). Providing this value will fix the pseudo-random sequence generated for the prototypeseed.aggregation(optional) is a list of aggregations to be performed on the outputs of your evaluation task.
Hook 🔗
Thehook keyword is used to save or display results generated during the execution of a workflow.
The generic way to use it is to write either hook(workDirectory / "path/of/a/file.csv") to save the results in a CSV file, or hook display to display the results in the standard output.
There are some arguments specific to the
Replication task which can be added to the hook:
outputis to choose what to do with the results as shown above, either a file path or the worddisplay,values = Seq(i, j)specifies which variables from the data flow should be saved or displayed, by default all variables from the dataflow are used,header = "Col1, Col2, ColZ"customises the header of the CSV file to be created with the string it receives as a parameter, please note that this only happens if the file doesn't exist when the hook is executed,arrayOnRow = trueforces the flattening of input lists such that all list variables are written to a single row/line of the CSV file, it defaults tofalse,includeSeedis aBooleanto specify whether you want the seed from theReplicationtask to be saved with the workflow variables,falseby default.
val mySeed = Val[Int]
val i = Val[Int]
val o = Val[Double]
val myModel =
ScalaTask("import scala.util.Random; val rng = new Random(mySeed); val o = i * 2 + 0.1 * rng.nextDouble()") set (
inputs += (i, mySeed),
outputs += (i, o)
)
Replication(
evaluation = myModel,
seed = mySeed,
sample = 100
) hook(
output = display,
values = Seq(i),
includeSeed = true)
For more details about hooks, check the corresponding Language page.